Tag: antibiotics

Explainable AI for Rational Antibiotic Discovery
Researchers now employ artificial intelligence (AI) models based on deep learning to make functional predictions about big datasets. While the concepts behind these networks are well established, their inner workings are often invisible to the user. The emerging area of explainable AI (xAI) provides model interpretation techniques that empower life science researchers to uncover the underlying basis on which AI models make such predictions.In this month’s episode, Deanna MacNeil from The Scientist spoke with Jim Collins from the Massachusetts Institute of Technology to learn how researchers are using explainable AI and artificial neural networks to gain mechanistic insights for large scale antibiotic discovery. Learn more
When an antibiotic fails: MIT scientists are using AI to target “sleeper” bacteria
Since the 1970s, modern antibiotic discovery has been experiencing a lull. Now the World Health Organization has declared the antimicrobial resistance crisis as one of the top 10 global public health threats.When an infection is treated repeatedly, clinicians run the risk of bacteria becoming resistant to the antibiotics. But why would an infection return after proper antibiotic treatment? One well-documented possibility is that the bacteria are becoming metabolically inert, escaping detection of traditional antibiotics that only respond to metabolic activity. When the danger has passed, the bacteria return to life and the infection reappears.
“Resistance is happening more over time, and recurring infections are due to this dormancy,” says Jackie Valeri, a former MIT-Takeda Fellow (centered within the MIT Abdul Latif Jameel Clinic for Machine Learning in Health) who recently earned her PhD in biological engineering from the Collins Lab. Valeri is the first author of a new paper published in this month’s print issue of Cell Chemical Biology that demonstrates how machine learning could help screen compounds that are lethal to dormant bacteria. Learn more
‘A landmark moment’: scientists use AI to design antibodies from scratch
Researchers have used generative artificial intelligence (AI) to help them make completely new antibodies for the first time.The proof-of-principle work, reported this week in a preprint on bioRxiv, raises the possibility of bringing AI-guided protein design to the therapeutic antibody market, which is worth hundreds of billions of dollars.
Antibodies — immune molecules that strongly attach to proteins implicated in disease — have conventionally been made using brute-force approaches that involve immunizing animals or screening vast numbers of molecules.
AI tools that can shortcut those costly efforts have the potential to “democratize the ability to design antibodies”, says study co-author Nathaniel Bennett, a computational biochemist at the University of Washington in Seattle. “Ten years from now, this is how we’re going to be designing antibodies.”
“It’s a really promising piece of research” that represents an important step in applying AI protein-design tools to making new antibodies, says Charlotte Deane, an immuno-informatician at the University of Oxford, UK. Learn more

Jim Collins: Discovery of the First New Structural Class of Antibiotics in Decades, Using A.I.
Jim Collins is one of the leading biomedical engineers in the world. He’s been elected to all 3 National Academies (Engineering, Science, and Medicine) and is one of the founders of the field of synthetic biology. In this conversation, we reviewed the seminal discoveries that he and his colleagues are making at the Antibiotics-AI Project at MIT. Learn more
A year of discovery
Why it matters: the world is in urgent need of new ideas and inspiration to address chronic and infectious diseases, climate change, energy demands and other complex and consequential problems.Here are some of the biggest discoveries and advances of 2023...
AI-assisted discovery: The push to use AI for science notched some advances in 2023 — including solving a famous math problem, finding a new class of antibiotics and predicting the structure of nearly 400,000 possible new materials, which are needed for next-generation batteries, solar cells and computing. Learn more

Using AI, MIT researchers identify a new class of antibiotic candidates
Using a type of artificial intelligence known as deep learning, MIT researchers have discovered a class of compounds that can kill a drug-resistant bacterium that causes more than 10,000 deaths in the United States every year. Learn more
New Class of Antibiotics Discovered Using AI
Antibiotic resistance is among the biggest global threats to human health. It was directly responsible for an estimated 1.27 million deaths in 2019 and contributed to nearly five million more. The problem only got worse during the COVID pandemic. And no new classes of antibiotics have been developed for decades. Learn more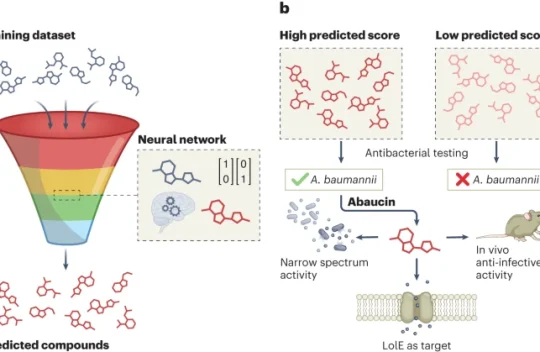
Antibiotic identified by AI
Computational approaches are emerging as powerful tools for the discovery of antibiotics. A study now uses machine learning to discover abaucin, a potent antibiotic that targets the bacterial pathogen Acinetobacter baumannii. Learn more
Using AI, scientists find a drug that could combat drug-resistant infections
The machine-learning algorithm identified a compound that kills Acinetobacter baumannii, a bacterium that lurks in many hospital settings. Learn more
