Tag: Jeremy Wohlwend

9 new breakthroughs in the fight against cancer
Lung cancer kills more people in the US yearly than the next three deadliest cancers combined. It's notoriously hard to detect the early stages of the disease with X-rays and scans alone. However, MIT scientists have developed an AI learning model to predict a person's likelihood of developing lung cancer up to six years in advance via a low-dose CT scan. Learn more
Promising new AI can detect early signs of lung cancer that doctors can’t see
Researchers in Boston are on the verge of what they say is a major advancement in lung cancer screening: Artificial intelligence that can detect early signs of the disease years before doctors would find it on a CT scan.The new AI tool, called Sybil, was developed by scientists at the Mass General Cancer Center and the Massachusetts Institute of Technology in Cambridge. In one study, it was shown to accurately predict whether a person will develop lung cancer in the next year 86% to 94% of the time. Learn more
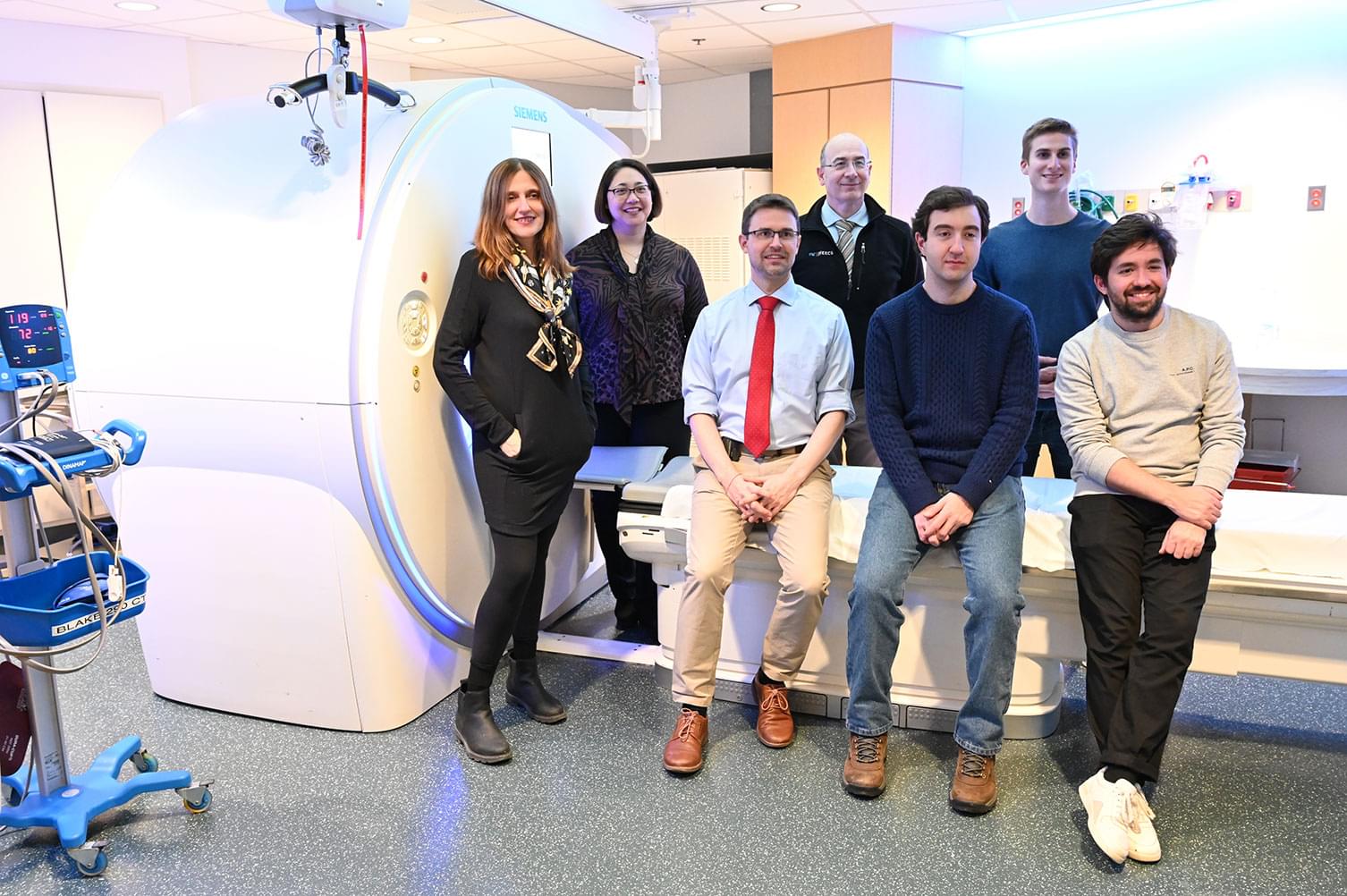
MIT researchers develop an AI model that can detect future lung cancer risk
The name Sybil has its origins in the oracles of Ancient Greece, also known as sibyls: feminine figures who were relied upon to relay divine knowledge of the unseen and the omnipotent past, present, and future. Now, the name has been excavated from antiquity and bestowed on an artificial intelligence tool for lung cancer risk assessment being developed by researchers at MIT's Abdul Latif Jameel Clinic for Machine Learning in Health, Mass General Cancer Center (MGCC), and Chang Gung Memorial Hospital (CGMH). Learn more
Remembering Octavian-Eugen Ganea, a gifted MIT postdoc AI researcher and beloved colleague
Octavian-Eugen Ganea, a gifted postdoctoral artificial intelligence researcher at the Abdul Latif Jameel Clinic for Machine Learning in Health (Jameel Clinic) and Computer Science and Artificial Intelligence Laboratory (CSAIL) passed away during a hike in French Polynesia on May 26. He was 34. Learn more

