Tag: drug discovery
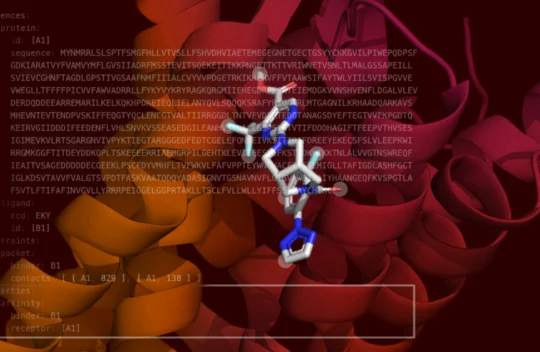
Boltz-2 Released to Democratize AI Molecular Modeling for Drug Discovery
Researchers from the Massachusetts Institute of Technology (MIT) Jameel Clinic for Machine Learning in Health have announced the open-source release of Boltz-2, which now predicts molecular binding affinity at newfound speed and accuracy to democratize commercial drug discovery. The model is available under the highly permissive MIT license, which allows commercial drug developers to use the model internally and apply their own proprietary data. Learn more