Tag: antibiotics
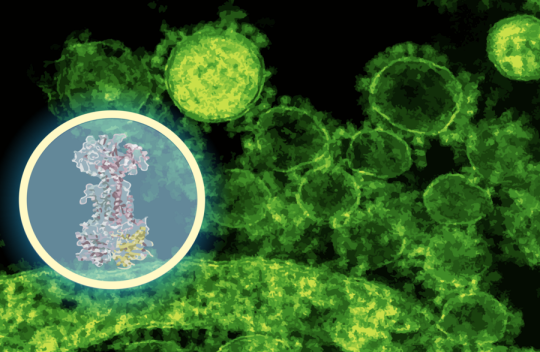
AI maps how a new antibiotic targets gut bacteria
“This discovery speaks to a central challenge in antibiotic development,” says Jon Stokes, senior author of a new paper on the work, assistant professor of biochemistry and biomedical sciences at McMaster, and research affiliate at MIT’s Abdul Latif Jameel Clinic for Machine Learning in Health. “The problem isn’t finding molecules that kill bacteria in a dish — we’ve been able to do that for a long time. A major hurdle is figuring out what those molecules actually do inside bacteria. Without that detailed understanding, you can’t develop these early-stage antibiotics into safe and effective therapies for patients.” Learn more
AI designs antibiotics for gonorrhoea and MRSA superbugs
Artificial intelligence has invented two new potential antibiotics that could kill drug-resistant gonorrhoea and MRSA, researchers have revealed.The drugs were designed atom-by-atom by the AI and killed the superbugs in laboratory and animal tests.
The two compounds still need years of refinement and clinical trials before they could be prescribed.
But the Massachusetts Institute of Technology (MIT) team behind it say AI could start a "second golden age" in antibiotic discovery. Learn more

AI used to design antibiotics that can combat drug-resistant superbugs gonorrhoea and MRSA
A team at Massachusetts Institute of Technology (MIT) used generative AI algorithms to design more than 36 million possible compounds. They also seemed to work in a new way - by disrupting bacterial cell membranes.Antibiotics kill bacteria, but some infections have become resistant to drugs.
It is estimated that drug-resistant bacterial infections cause nearly five million deaths per year worldwide.
Two compounds were found to be effective against gonorrhoea and MRSA infections - namely NG1 and DN1, respectively. Learn more

Using generative AI, researchers design compounds that can kill drug-resistant bacteria
With help from artificial intelligence, MIT researchers have designed novel antibiotics that can combat two hard-to-treat infections: drug-resistant Neisseria gonorrhoeae and multi-drug-resistant Staphylococcus aureus (MRSA).Using generative AI algorithms, the research team designed more than 36 million possible compounds and computationally screened them for antimicrobial properties. The top candidates they discovered are structurally distinct from any existing antibiotics, and they appear to work by novel mechanisms that disrupt bacterial cell membranes.
This approach allowed the researchers to generate and evaluate theoretical compounds that have never been seen before — a strategy that they now hope to apply to identify and design compounds with activity against other species of bacteria.
“We’re excited about the new possibilities that this project opens up for antibiotics development. Our work shows the power of AI from a drug design standpoint, and enables us to exploit much larger chemical spaces that were previously inaccessible,” says James Collins, the Termeer Professor of Medical Engineering and Science in MIT’s Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering. Learn more

AI Impact Awards 2025: How 7 Health Care Winners Measure Impact
In 2020, researchers at the Collins Lab at MIT made a landmark discovery when they used AI to identify a new class of antibiotics. Phare Bio was born from that breakthrough, and has since leveraged AI to uncover two additional novel antibiotic classes.The company's model prioritizes the superbugs identified as the most dangerous by the CDC and the WHO, and predicts drug efficacy, toxicity and pharmacokinetics with high accuracy. Phare Bio has also developed AIBiotics, a generative AI platform that designs new antibiotics.
Ultimately, the company aims to improve the efficiency of antibiotic research and development, according to Dr. Akhila Kosaraju, its president and CEO.
How does it measure that? Ultimately, by "taking better and fewer shots on goal," Kosaraju told Newsweek. It often costs between $1.3 and $1.5 billion to get a single drug over the finish line for FDA approval.
"Those numbers are so high [because they] encompass all of the failures along the way to get to that one exceptional drug," Kosaraju said. "If we can reduce the number of shots on goal substantially, we can half or quarter the cost and time to get these drugs into clinical trials, and then ultimately to be FDA-approved."
To see the full list of AI Impact winners, visit the official page for Newsweek's AI Impact Awards. Learn more

ARPA-H project to accelerate the discovery and development of new antibiotics using generative AI
Today, the U.S. Department of Health and Human Services (HHS) through the Advanced Research Projects Agency for Health (ARPA-H) announced funding for the Transforming Antibiotic R&D with Generative AI to stop Emerging Threats (TARGET) project, which will use AI to speed the discovery and development of new classes of antibiotics. This program is another action to support the United States’ longstanding commitment to combating antimicrobial resistance (AMR), from groundbreaking innovation to international collaboration. The U.S. is a global leader in the fight against AMR and has a demonstrated track record of progress in protecting people, animals, and the environment from the threat of AMR domestically and globally.“Antibiotic resistance is a real and urgent threat affecting millions of people. We need to prevent infections and conserve the antibiotics we have. We also urgently need new drugs to treat these increasingly resistant infections. This project will use AI to speed this needed innovation and help ensure we have the medicines we need to keep people alive,” said Secretary Xavier Becerra. Learn more

How Machines Learned to Discover Drugs
When I first became a doctor, I cared for an older man whom I’ll call Ted. He was so sick with pneumonia that he was struggling to breathe. His primary-care physician had prescribed one antibiotic after another, but his symptoms had only worsened; by the time I saw him in the hospital, he had a high fever and was coughing up blood. His lungs seemed to be infected with methicillin-resistant Staphylococcus aureus (MRSA), a bacterium so hardy that few drugs can kill it. I placed an oxygen tube in his nostrils, and one of my colleagues inserted an I.V. into his arm. We decided to give him vancomycin, a last line of defense against otherwise untreatable infections.Ted recovered with astonishing speed. When I stopped by the next morning, he smiled and removed the oxygen tube, letting it dangle near his neck like a pendant. Then he pointed to the I.V. pole near his bed, where a clear liquid was dripping from a bag and into his veins.
“Where did that stuff come from?” Ted asked.
“The pharmacy,” I said.
“No, I mean, where did it come from?”
At the time, I could barely pronounce the names of medications, let alone hold forth on their provenance. “I’ll have to get back to you,” I told Ted. He was discharged before I could. But, in the years that followed, I often thought about his question. Every day, I administer medicines whose origins are a mystery to me. I occasionally meet a patient for whom I have no effective treatment to offer, and Ted’s inquiry starts to seem existential. Where do drugs come from, and how can we get more of them? Learn more

Explainable AI for Rational Antibiotic Discovery
Researchers now employ artificial intelligence (AI) models based on deep learning to make functional predictions about big datasets. While the concepts behind these networks are well established, their inner workings are often invisible to the user. The emerging area of explainable AI (xAI) provides model interpretation techniques that empower life science researchers to uncover the underlying basis on which AI models make such predictions.In this month’s episode, Deanna MacNeil from The Scientist spoke with Jim Collins from the Massachusetts Institute of Technology to learn how researchers are using explainable AI and artificial neural networks to gain mechanistic insights for large scale antibiotic discovery. Learn more
When an antibiotic fails: MIT scientists are using AI to target “sleeper” bacteria
Since the 1970s, modern antibiotic discovery has been experiencing a lull. Now the World Health Organization has declared the antimicrobial resistance crisis as one of the top 10 global public health threats.When an infection is treated repeatedly, clinicians run the risk of bacteria becoming resistant to the antibiotics. But why would an infection return after proper antibiotic treatment? One well-documented possibility is that the bacteria are becoming metabolically inert, escaping detection of traditional antibiotics that only respond to metabolic activity. When the danger has passed, the bacteria return to life and the infection reappears.
“Resistance is happening more over time, and recurring infections are due to this dormancy,” says Jackie Valeri, a former MIT-Takeda Fellow (centered within the MIT Abdul Latif Jameel Clinic for Machine Learning in Health) who recently earned her PhD in biological engineering from the Collins Lab. Valeri is the first author of a new paper published in this month’s print issue of Cell Chemical Biology that demonstrates how machine learning could help screen compounds that are lethal to dormant bacteria. Learn more


